Biodiversity metrics and metabarcoding
2/2/24
Summary
- The MetabarSchool Package
- What do the reading numbers per PCR mean?
- Rarefaction vs. relative frequencies
- alpha diversity metrics
- beta diversity metrics
- multidimentionnal analysis
- comparison between datasets
The MetabarSchool Package
Installing the package
You need the devtools package
Then you can install MetabarSchool
You will also need the vegan package
The dataset
The mock community
A 16 plants mock community
| species | taxid | Relative aboundance | |
|---|---|---|---|
| 1 | Taxus baccata | 25629 | 1/2 |
| 2 | Salvia pratensis | 49216 | 1/4 |
| 3 | Populus tremula | 113636 | 1/8 |
| 4 | Rumex acetosa | 41241 | 1/16 |
| 5 | Carpinus betulus | 12990 | 1/32 |
| 6 | Fraxinus excelsior | 38873 | 1/64 |
| 7 | Picea abies | 3329 | 1/128 |
| 8 | Lonicera xylosteum | 439142 | 1/256 |
| 9 | Abies alba | 45372 | 1/512 |
| 10 | Acer campestre | 66205 | 1/1024 |
| 11 | Briza media | 281077 | 1/2048 |
| 12 | Rosa canina | 74635 | 1/4096 |
| 13 | Capsella bursa-pastoris | 3719 | 1/8192 |
| 14 | Geranium robertianum | 122183 | 1/16384 |
| 15 | Rhododendron ferrugineum | 49622 | 1/32768 |
| 16 | Lotus corniculatus | 47247 | 1/65536 |
The experiment
192 PCR of the mock community using SPER02 trnL-P6-Loop primers
6 dilutions of the mock community: 1/1, 1/2, 1/4, 1/8, 1/16, 1/32
32 repeats per dilution
Loading data
positive.countread count matrix \(192 \; PCRs \; \times \; 24330 \; MOTUs\)
| P000001 | P000002 | P000003 | P000004 | P000005 | |
|---|---|---|---|---|---|
| sample.TM_POS_d16_1_a_A1 | 1167 | 4477 | 779 | 0 | 12 |
| sample.TM_POS_d16_1_a_B1 | 1072 | 5077 | 985 | 2 | 8 |
| sample.TM_POS_d16_1_b_A2 | 919 | 3599 | 601 | 0 | 10 |
| sample.TM_POS_d16_1_b_B2 | 704 | 4129 | 835 | 2 | 15 |
| sample.TM_POS_d16_2_a_A1 | 1155 | 5341 | 1023 | 2 | 6 |
Loading data
positive.samplesa 192 rowsdata.frameof 2 columns describing each PCR
| dilution | repeats | |
|---|---|---|
| sample.TM_POS_d16_1_a_A1 | 2 | 1.a.A1 |
| sample.TM_POS_d16_1_a_B1 | 2 | 1.a.B1 |
| sample.TM_POS_d16_1_b_A2 | 2 | 1.b.A2 |
Loading data
positive.motus: a 24330 rowsdata.frameof 4 columns describing each MOTU
| dilution | species | taxid | true | |
|---|---|---|---|---|
| P000001 | 0.250 | Salvia pratensis | 49216 | TRUE |
| P000002 | 0.125 | Populus tremula | 113636 | TRUE |
| P000003 | 0.500 | Taxus baccata | 25629 | TRUE |
Removing singleton sequences
Singleton sequences are observed only once over the complete dataset.
| FALSE | TRUE |
|---|---|
| 5579 | 18751 |
We discard them they are unanimously considered as rubbish.
positive.countis now a \(192 \; PCRs \; \times \; 5579 \; MOTUs\) matrix
Not all the PCR have the same number of reads
Despite all standardization efforts

Is it related to the amount of DNA in the extract ?
What do the reading numbers per PCR mean?
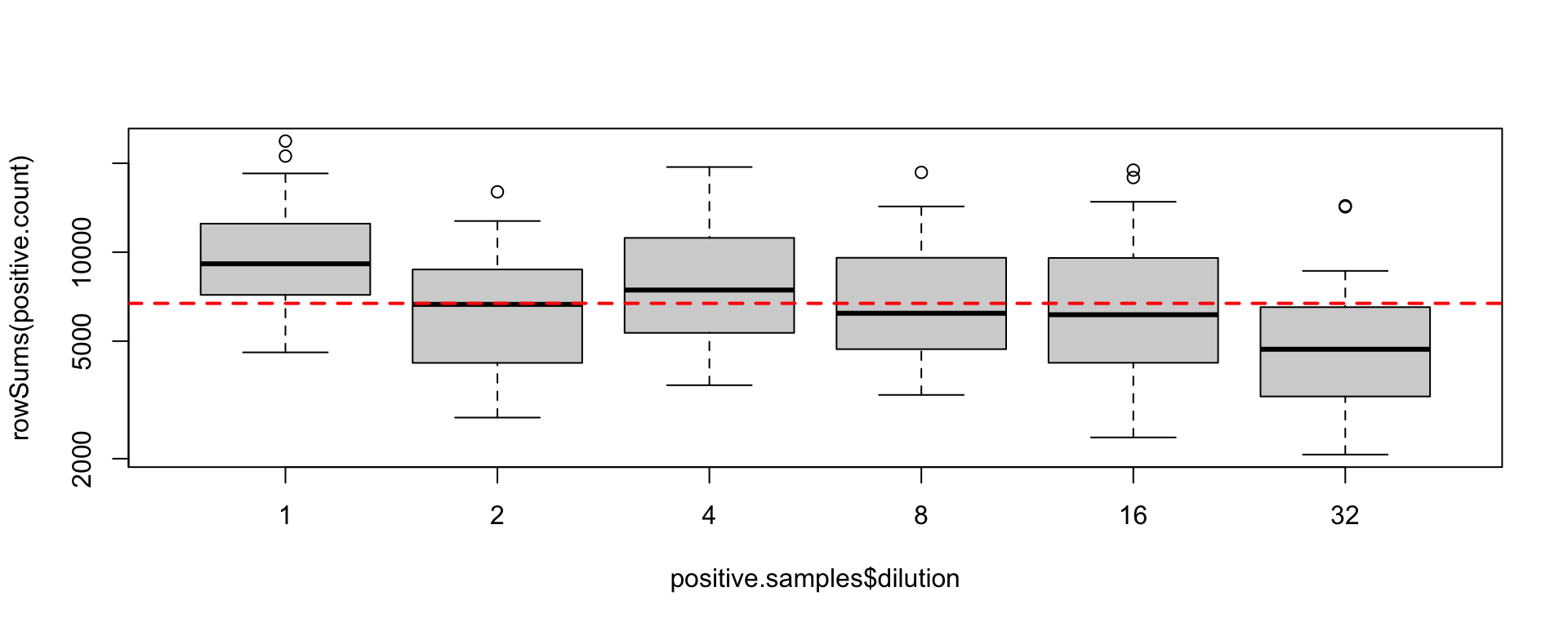
You must normalize your read counts
Two options:
Rarefaction
Randomly subsample the same number of reads for all the PCRs
Relative frequencies
Divide the read count of each MOTU in each sample by the total total read count of the same sample
\[ \text{Relative fequency}(Motu_i,Sample_j) = \frac{\text{Read count}(Motu_i,Sample_j)}{\sum_{k=1}^n\text{Read count}(Motu_k,Sample_j)} \]
Rarefying read count (1)
- We look for the minimum read number per PCR
Rarefying read count (2)
Rarefying read count (3)
Identifying the MOTUs with reads count greater than \(0\) after rarefaction.
P000001 P000002 P000003 P000004 P000005
TRUE TRUE TRUE TRUE TRUE Rarefying read count (4)

The MOTUs removed by rarefaction were at most occurring 20 times
The MOTUs kept by rarefaction were at least occurring 2 times
Rarefying read count (5)
Keep only sequences with reads after rarefaction
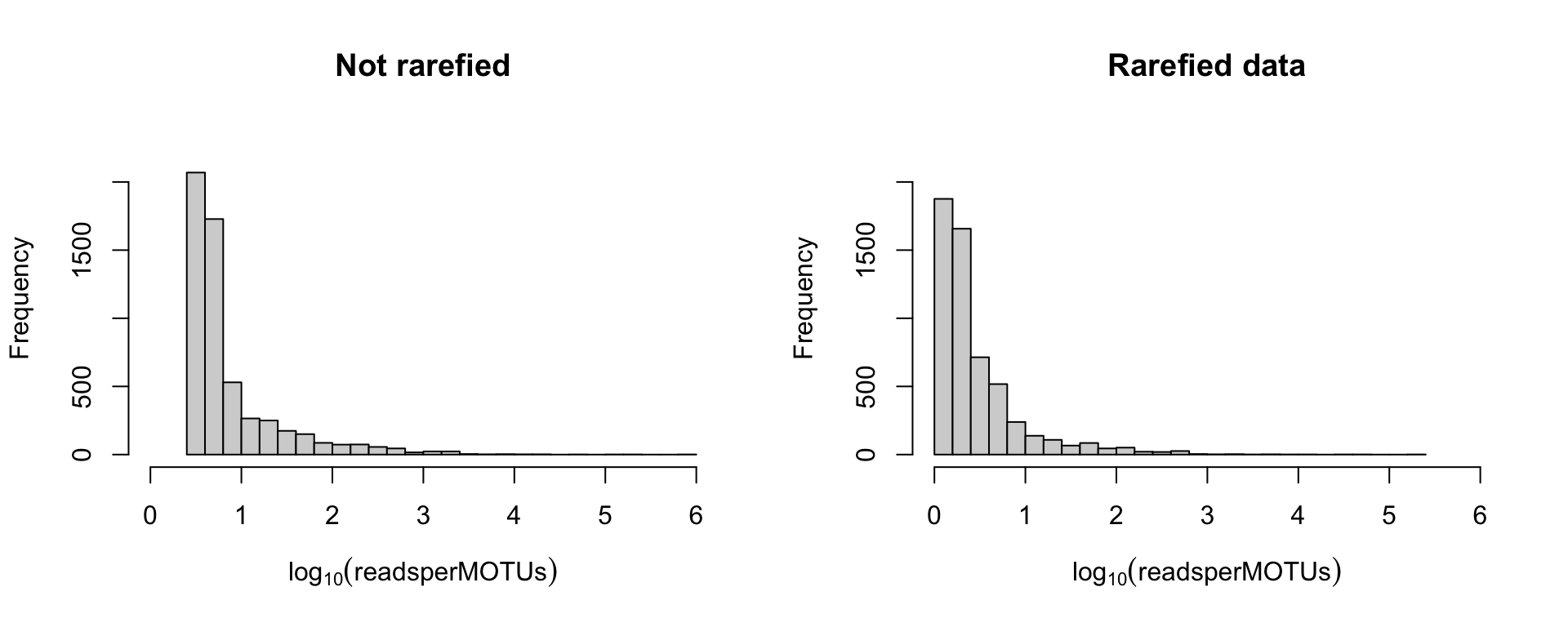
Why rarefying ?
Increasing the number of reads just increase the description of the subpart of the PCR you have sequenced.
Transforming read counts to relative frequencies
No sequences will be set to zero
Measuring diversity
The different types of diversity
Whittaker (2010)
\(\alpha\text{-diversity}\) : Mean diversity per site (\(species/site\))
\(\gamma\text{-diversity}\) : Regional biodiversity (\(species/region\))
\(\beta\text{-diversity}\) : \(\beta = \frac{\gamma}{\alpha}\) (\(sites/region\))
\(\alpha\)-diversity
Which is th most diverse environment ?
| A | B | C | D | E | F | G | |
|---|---|---|---|---|---|---|---|
| Environment.1 | 0.25 | 0.25 | 0.25 | 0.25 | 0.00 | 0.00 | 0.00 |
| Environment.2 | 0.55 | 0.07 | 0.02 | 0.17 | 0.07 | 0.07 | 0.03 |
Richness
The actual number of species present in your environement whatever their aboundances
| S | |
|---|---|
| Environment.1 | 4 |
| Environment.2 | 7 |
Gini-Simpson’s index
The Simpson’s index is the probability of having the same species twice when you randomly select two specimens.
\[
\lambda =\sum _{i=1}^{S}p_{i}^{2}
\]
\(\lambda\) decrease when complexity of your ecosystem increase.
Gini-Simpson’s index defined as \(1-\lambda\) increase with diversity
| Gini.Simpson | |
|---|---|
| Environment.1 | 0.7500 |
| Environment.2 | 0.6526 |
Shannon entropy
Shannon entropy is based on information theory:
if \(A\) is a community where every species are equally represented then \[ H(A) = \log|A| \]
| Shannon.index | |
|---|---|
| Environment.1 | 1.386294 |
| Environment.2 | 1.371925 |
Hill’s number
As : \[
H(A) = \log|A| \;\Rightarrow\; ^1D = e^{H(A)}
\]
where \(^1D\) is the theoretical number of species in a evenly distributed community that would have the same Shannon’s entropy than ours.
| Hill.Numbers | |
|---|---|
| Environment.1 | 4.000000 |
| Environment.2 | 3.942933 |
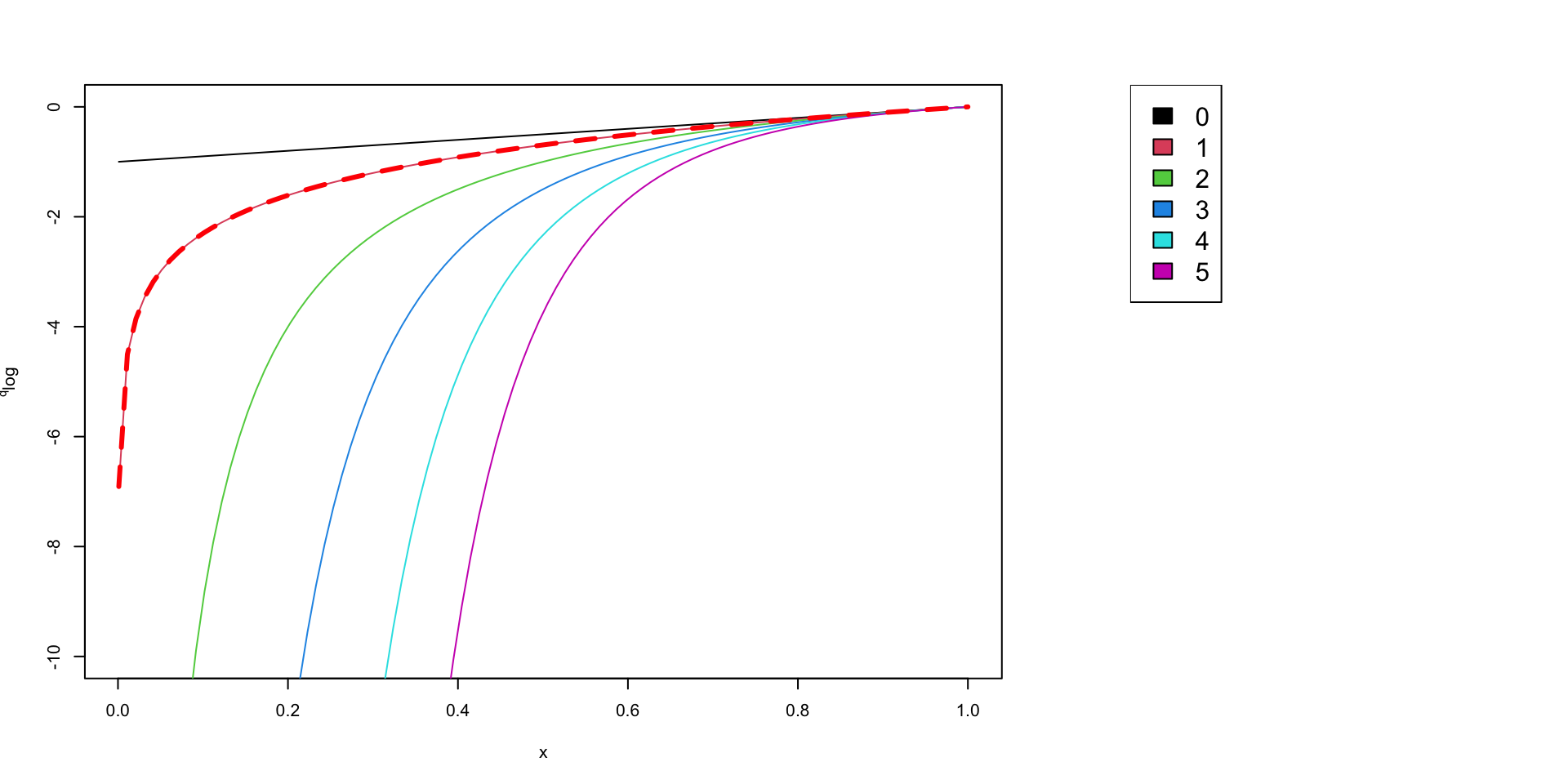
Generalized logaritmic function
Based on the generalized entropy Tsallis (1994) we can propose a generalized form of logarithm.
\[ ^q\log(x) = \frac{x^{(1-q)}-1}{1-q} \]
The function is not defined for \(q=1\) but when \(q \longrightarrow 1\;,\; ^q\log(x) \longrightarrow \log(x)\)
\[ ^q\log(x) = \left\{ \begin{align} \log(x),& \text{if } q = 1\\ \frac{x^{(1-q)}-1}{1-q},& \text{otherwise} \end{align} \right. \]
Impact of \(q\) on the log_q function
And its inverse function
\[ ^qe^x = \left\{ \begin{align} e^x,& \text{if } x = 1 \\ (1 + x(1-q))^{(\frac{1}{1-q})},& \text{otherwise} \end{align} \right. \]
Generalised Shannon entropy
\[ ^qH = - \sum_{i=1}^S p_i \; ^q\log p_i \]
and generalized the previously presented Hill’s number
\[ ^qD=^qe^{^qH} \]
Biodiversity spectrum (1)
Biodiversity spectrum (2)
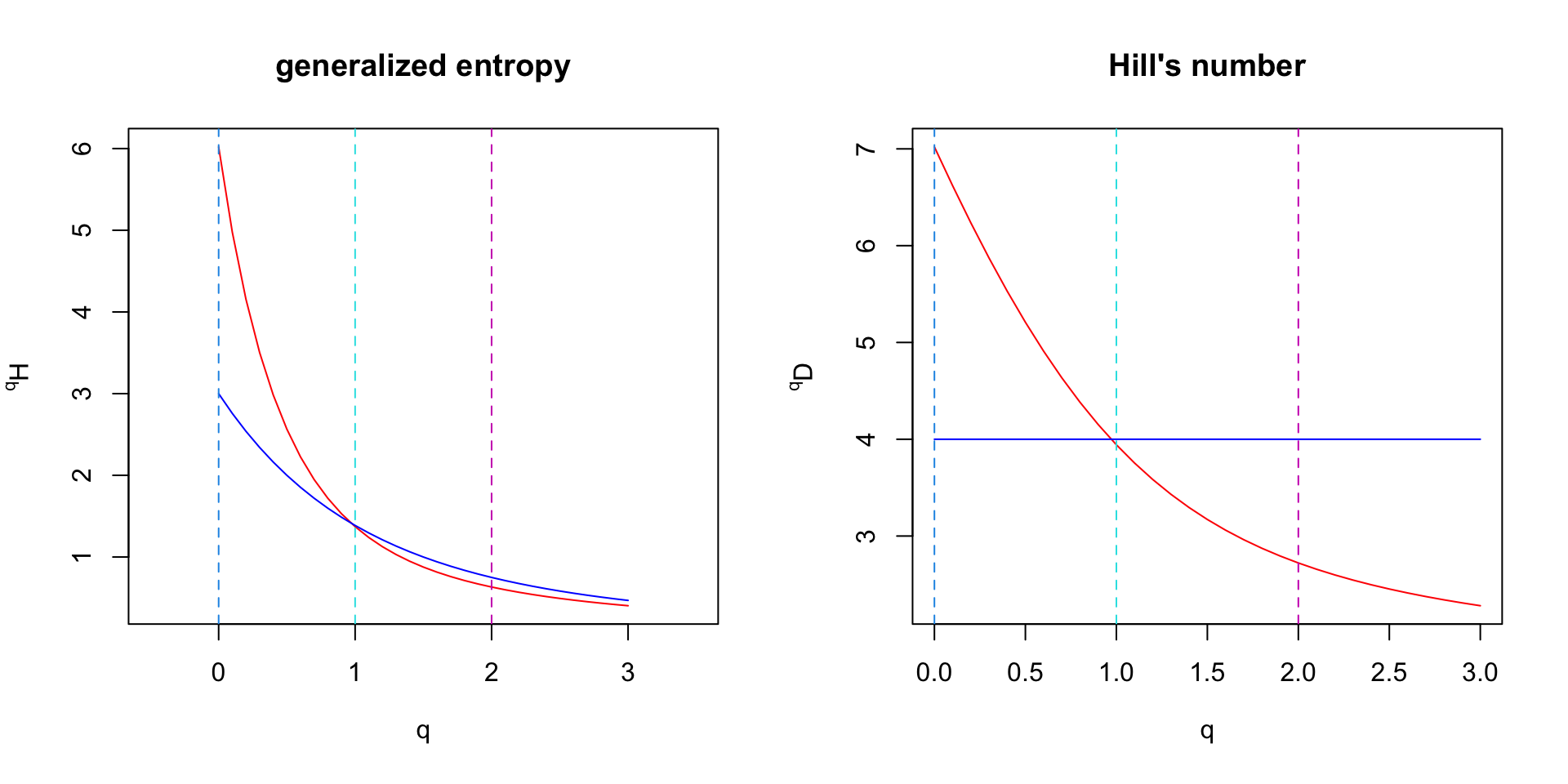
Generalized entropy \(vs\) \(\alpha\)-diversity indices
\(^0H(X) = S - 1\) : the richness minus one.
\(^1H(X) = H^{\prime}\) : the Shannon’s entropy.
\(^2H(X) = 1 - \lambda\) : Gini-Simpson’s index.
When computing the exponential of entropy : Hill’s number
\(^0D(X) = S\) : The richness.
\(^1D(X) = e^{H^{\prime}}\) : The number of species in an even community having the same \(H^{\prime}\).
\(^2D(X) = 1 / \lambda\) : The number of species in an even community having the same Gini-Simpson’s index.
\(q\) can be considered as a penality you give to rare species
when \(q=0\) all the species have the same weight
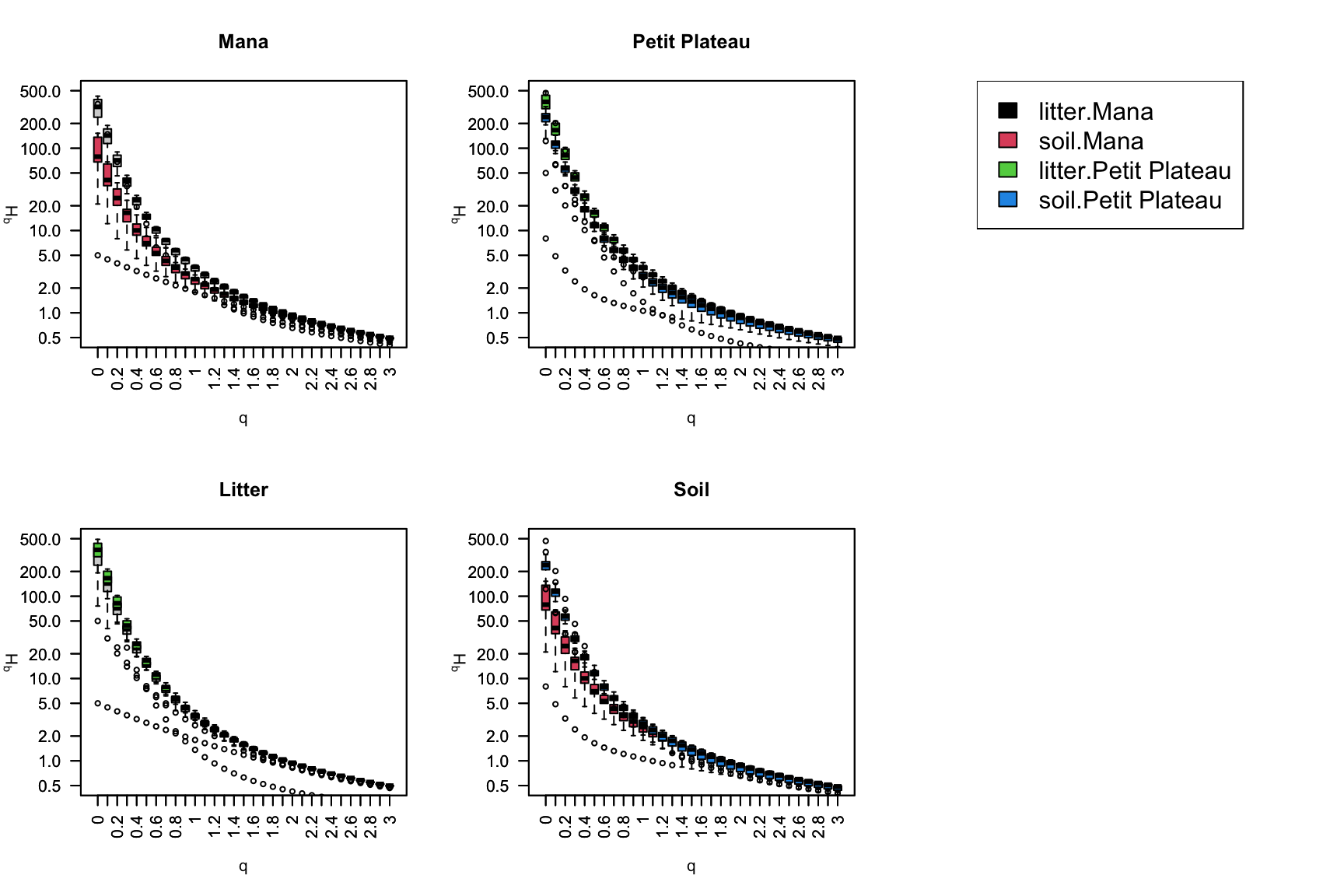
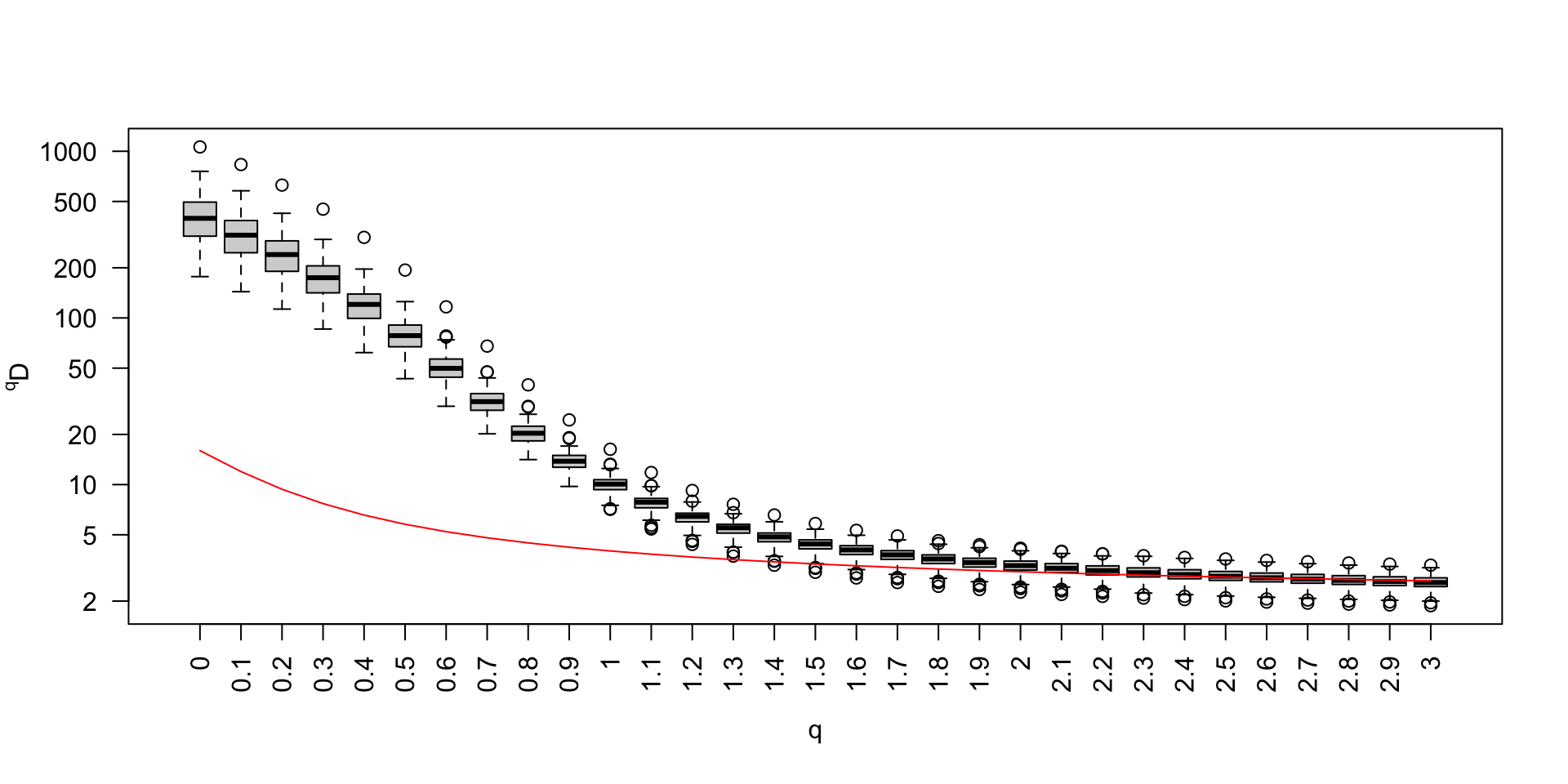
Biodiversity spectrum of the mock community
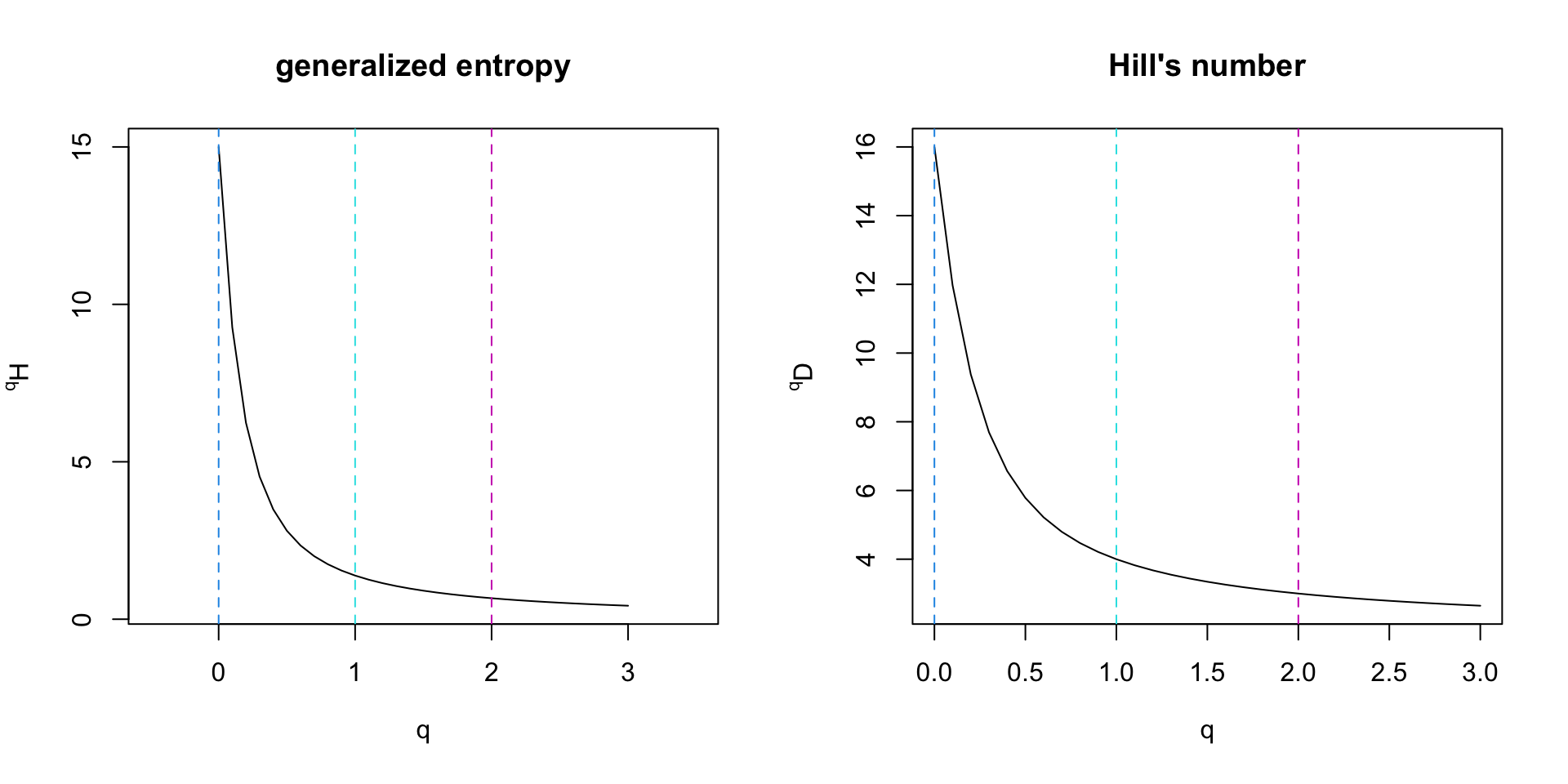
Biodiversity spectrum and metabarcoding (1)
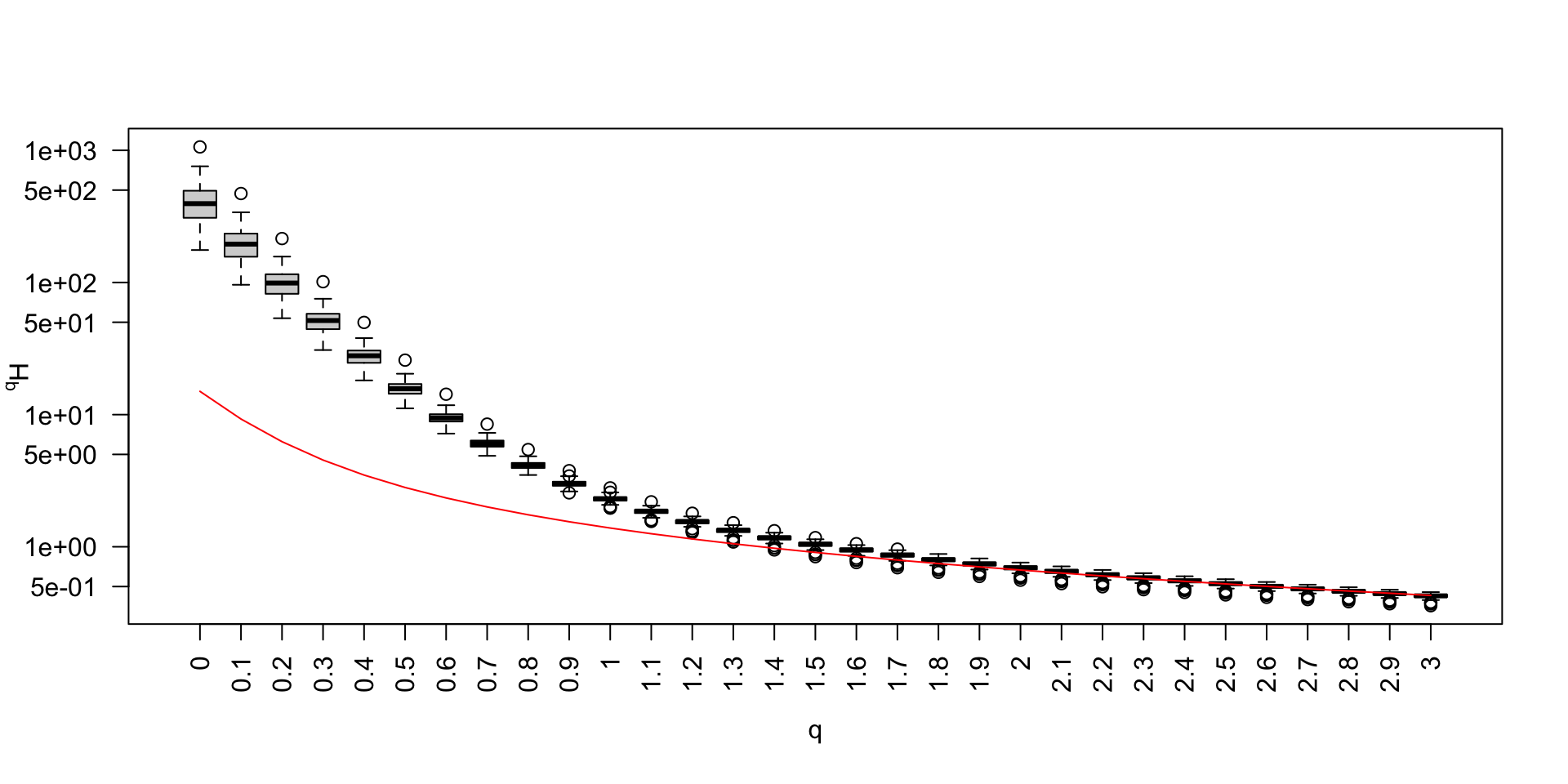
Biodiversity spectrum and metabarcoding (2)
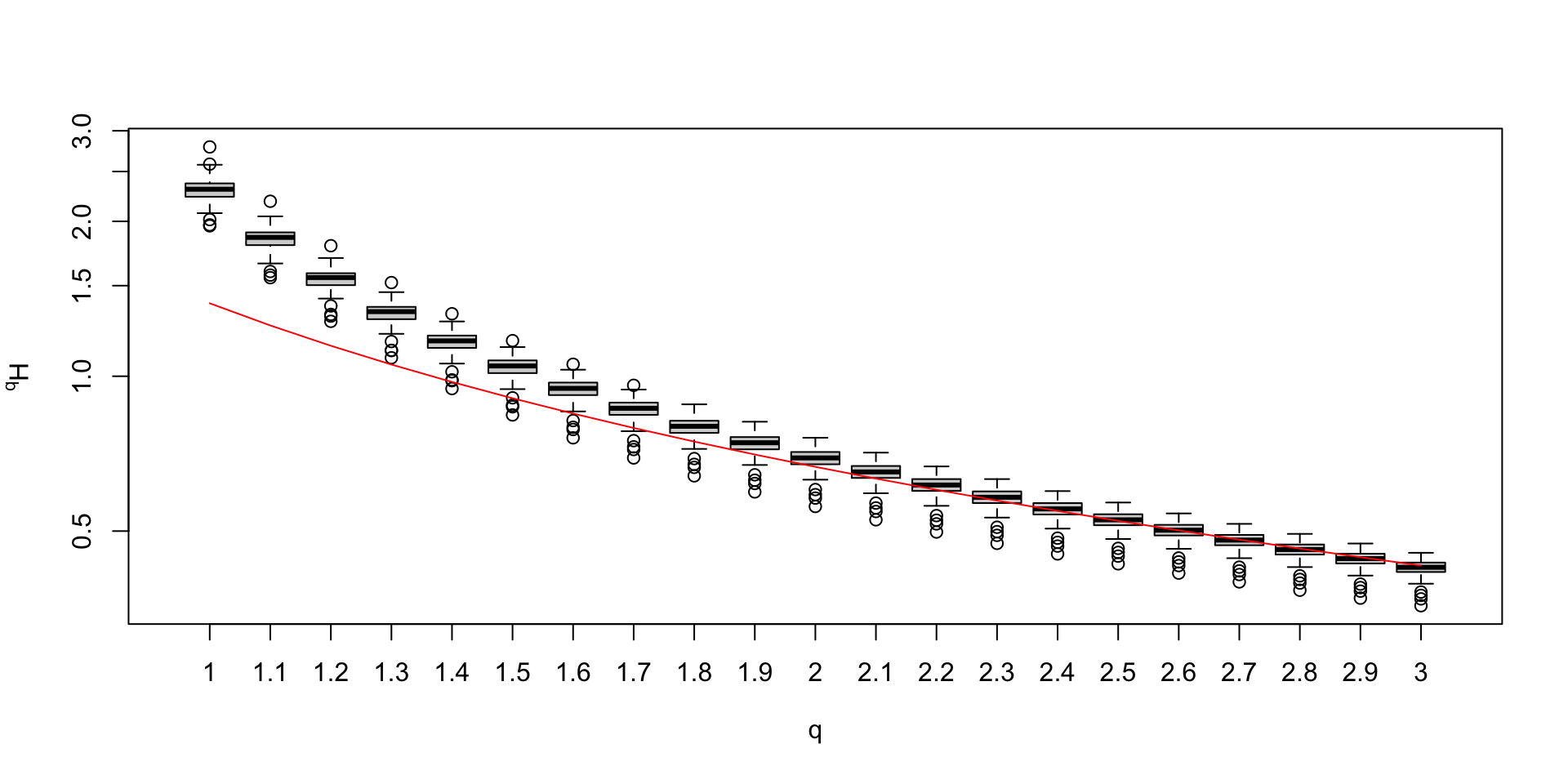
Biodiversity spectrum and metabarcoding (3)
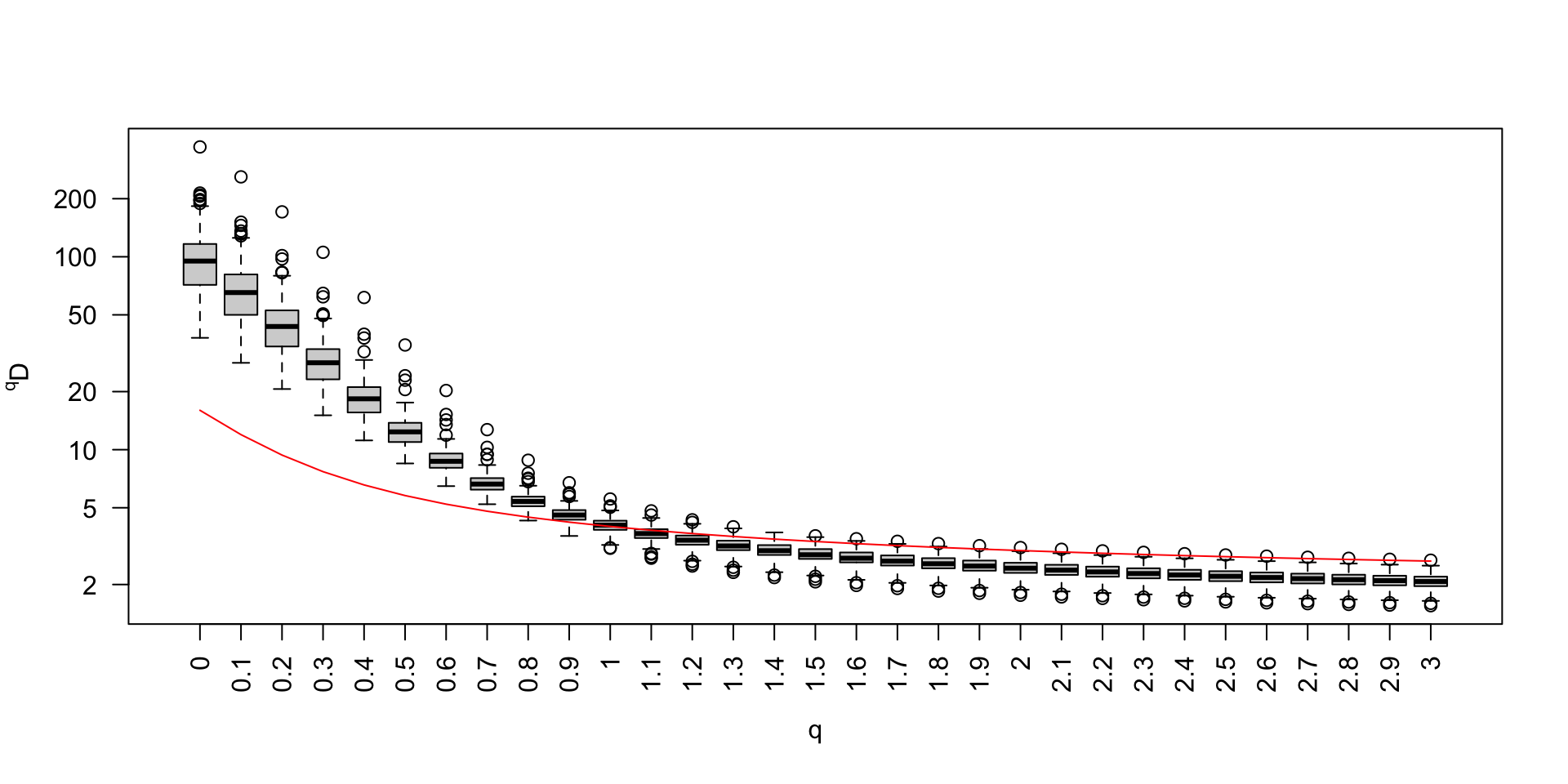
Impact of data cleaning on \(\alpha\)-diversity (1)
We realize a basic cleaning:
- removing signletons
- too short or long sequences
- clustering data using
obiclean
obigrep -p 'count > 1' \
positifs.uniq.annotated.fasta \
> positifs.uniq.annotated.no.singleton.fasta
obigrep -l 10 -L 150 \
positifs.uniq.annotated.no.singleton.fasta \
> positifs.uniq.annotated.good.length.fasta
obiclean -s merged_sample -H -C -r 0.1 \
positifs.uniq.annotated.good.length.fasta \
> positifs.uniq.annotated.clean.fastaImpact of data cleaning on \(\alpha\)-diversity (2)

Impact of data cleaning on \(\alpha\)-diversity (3)
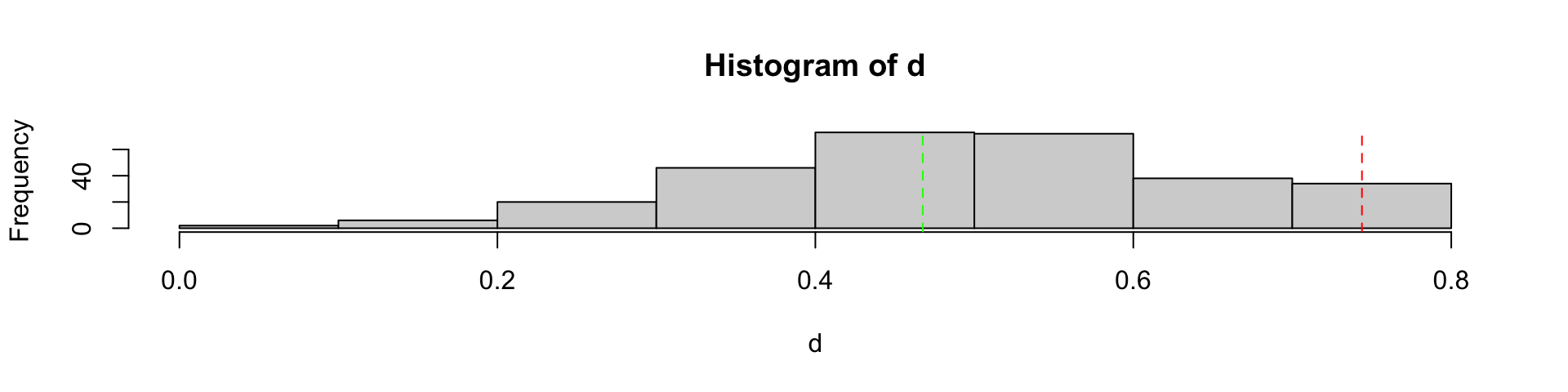
\(\beta\)-diversity
Dissimilarity indices or non-metric distances
of how far apart objects \(A\) and \(B\) are.
Properties
\[ \begin{align} d(A,B) \geqslant& 0 \\ d(A,B) =& d(B,A) \\ d(A,B) =& 0 \iff A = B \\ \end{align} \]
Some dissimilarity indices
Bray-Curtis
Relying on contengency table (quantitative data)
\[ {\displaystyle BC(A,B)=1-{\frac {2\sum _{i=1}^{p}min(N_{Ai},N_{Bi})}{\sum _{i=1}^{p}(N_{Ai}+N_{Bi})}}}, \; \text{with }p\text{ the total number of species} \]
Jaccard indices
Relying on presence absence data
\[ J(A,B) = {{|A \cap B|}\over{|A \cup B|}} = {{|A \cap B|}\over{|A| + |B| - |A \cap B|}}. \]
Metrics or distances
A metric is a dissimilarity index verifying the subadditivity also named triangle inequality
\[ \begin{align} d(A,B) \geqslant& 0 \\ d(A,B) =& \;d(B,A) \\ d(A,B) =& \;0 \iff A = B \\ d(A,B) \leqslant& \;d(A,C) + d(C,B) \end{align} \]
Some metrics
Computing
\[ \begin{align} d_e =& \sqrt{(x_A - x_B)^2 + (y_A - y_B)^2} \\ d_m =& |x_A - x_B| + |y_A - y_B| \\ d_c =& \max(|x_A - x_B| , |y_A - y_B|) \\ \end{align} \]
Generalizable on a n-dimension space
Considering 2 points \(A\) and \(B\) defined by \(n\) variables
\[ \begin{align} A :& (a_1,a_2,a_3,...,a_n) \\ B :& (b_1,b_2,b_3,...,b_n) \end{align} \]
with \(a_i\) and \(b_i\) being respectively the value of the \(i^{th}\) variable for \(A\) and \(B\).
\[ \begin{align} d_e =& \sqrt{\sum_{i=1}^{n}(a_i - b_i)^2 } \\ d_m =& \sum_{i=1}^{n}\left| a_i - b_i \right| \\ d_c =& \max\limits_{1\leqslant i \leqslant n}\left|a_i - b_i\right| \\ \end{align} \]
For the fun… ;-)
You can generalize those distances as a norm of order \(k\)
\[ d^k = \sqrt[k]{\sum_{i=1}^n|a_i - b_i|^k} \]
- \(k=1 \Rightarrow D_m\) Manhatan distance
- \(k=2 \Rightarrow D_e\) Euclidean distance
- \(k=\infty \Rightarrow D_c\) Chebychev distance
Metrics and ultrametrics
Metric
\[ d(x,z)\leqslant d(x,y)+d(y,z) \]
Ultrametric
\[ d(x,z)\leq \max(d(x,y),d(y,z)) \]
Why it is nice to use metrics ?
- A metric induce a metric space
- In a metric space rotations are isometries
- This means that rotations are not changing distances between objects
- Multidimensional scaling (PCA, PCoA, CoA…) are rotations
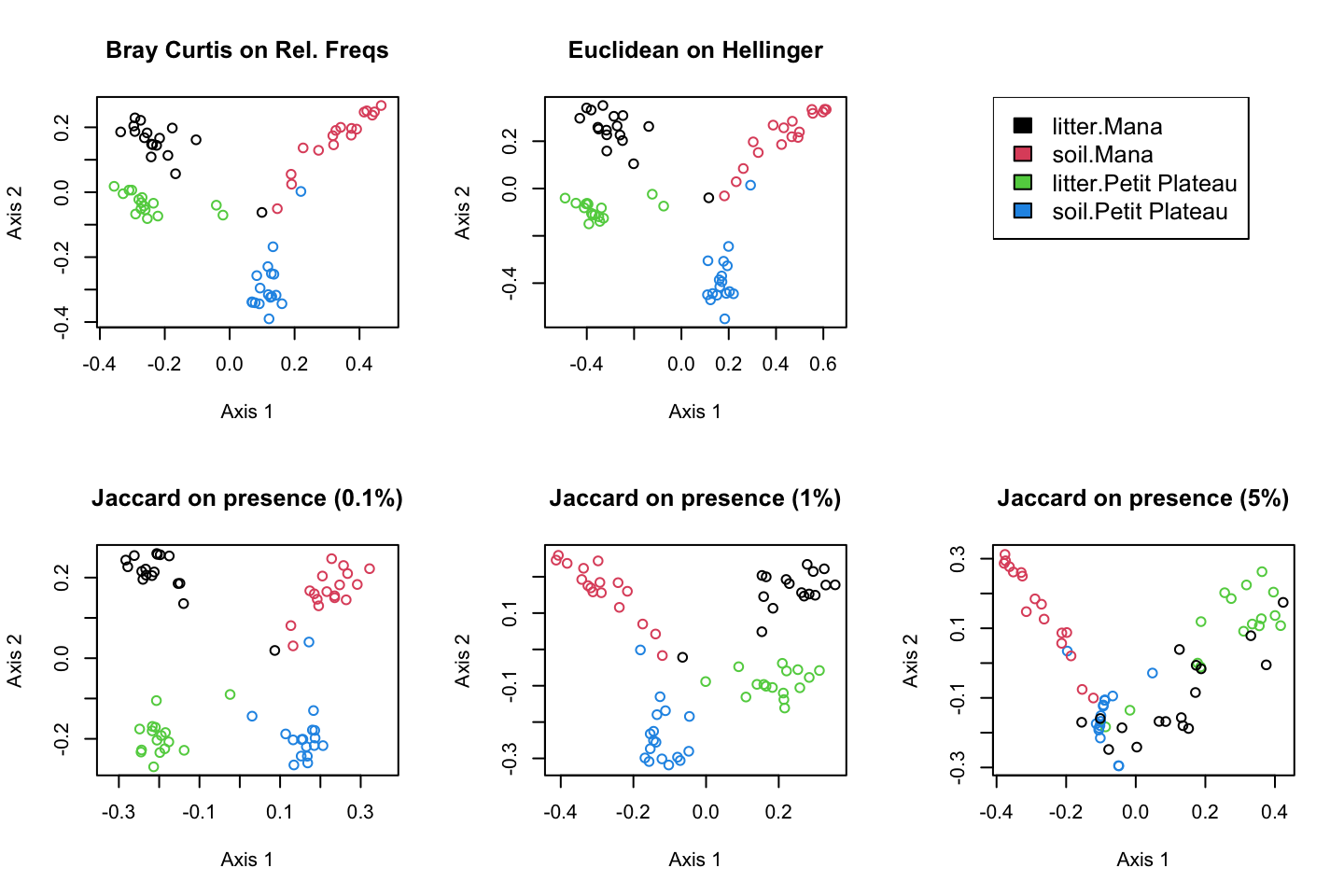
The data set
We analyzed two forest sites in French Guiana
Mana : Soil is composed of white sands.
Petit Plateau : Terra firme (firm land). In the Amazon, it corresponds to the area of the forest that is not flooded during high water periods. The terra firme is characterized by old and poor soils.
At each site, twice sixteen samples where collected over an hectar
Sixteen samples of soil. Each of them is constituted by a mix of five cores of 50g from the 10 first centimeters of soil covering half square meter.
Sixteen samples of litter. Each of them is constituted by the total litter collecter over the same half square meter where soil was sampled
Clean out bad PCR cycle 1
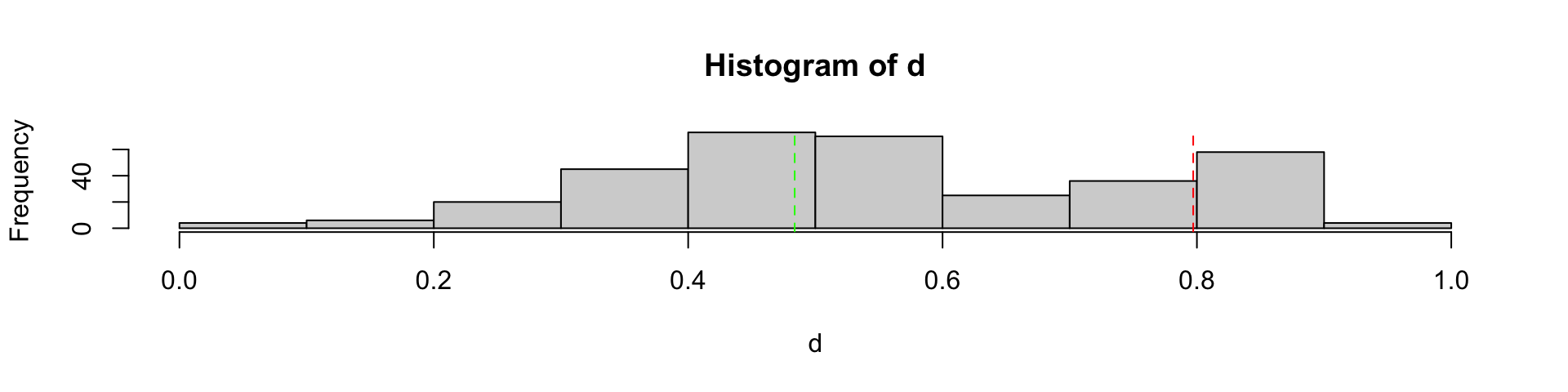
Clean out bad PCR cycle 2
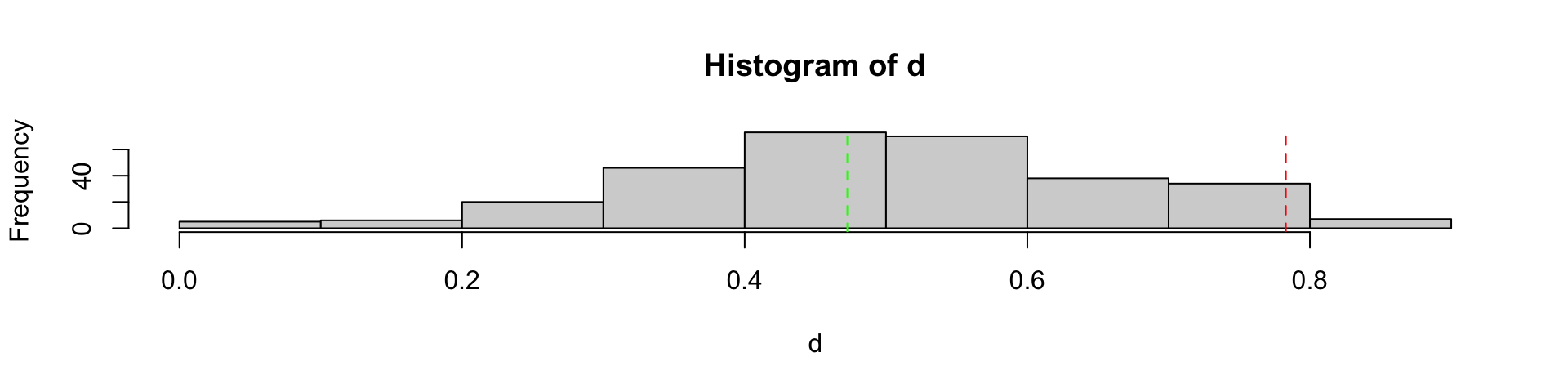
Clean out bad PCR cycle 3
Averaging good PCR replicates (1)
guiana.samples.clean = cbind(guiana.samples.clean,s[rownames(guiana.samples.clean),])
guiana.count.mean = aggregate(decostand(guiana.count.clean,method = "total"),
by = list(guiana.samples.clean$sample),
FUN=mean)
n = guiana.count.mean[,1]
guiana.count.mean = guiana.count.mean[,-1]
rownames(guiana.count.mean)=as.character(n)
guiana.count.mean = as.matrix(guiana.count.mean)
dim(guiana.count.mean)[1] 84 7884Averaging good PCR replicates (2)
guiana.samples.mean = aggregate(guiana.samples.clean,
by = list(guiana.samples.clean$sample),
FUN=function(i) i[1])
n = guiana.samples.mean[,1]
guiana.samples.mean = guiana.samples.mean[,-1]
rownames(guiana.samples.mean)=as.character(n)
dim(guiana.samples.mean)[1] 84 17Keep only samples
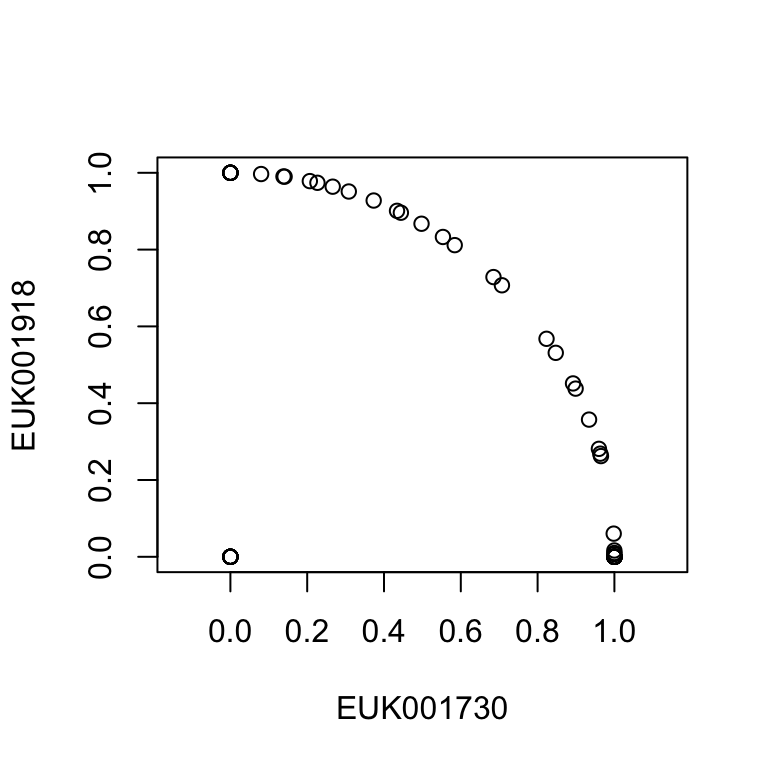
Estimating similarity between samples
guiana.hellinger.final = decostand(guiana.count.final,method = "hellinger")
guiana.relfreq.final = decostand(guiana.count.final,method = "total")
guiana.presence.1.final = guiana.relfreq.final > 0.001
guiana.presence.10.final = guiana.relfreq.final > 0.01
guiana.presence.50.final = guiana.relfreq.final > 0.05
guiana.bc.dist = vegdist(guiana.relfreq.final,method = "bray")
guiana.euc.dist = vegdist(guiana.hellinger.final,method = "euclidean")
guiana.jac.1.dist = vegdist(guiana.presence.1.final,method = "jaccard")
guiana.jac.10.dist = vegdist(guiana.presence.10.final,method = "jaccard")
guiana.jac.50.dist = vegdist(guiana.presence.50.final,method = "jaccard")Euclidean distance on Hellinger transformation
Bray-Curtis distance on relative frequencies
\[ BC_{jk}=1-{\frac {2\sum _{i=1}^{p}min(N_{ij},N_{ik})}{\sum _{i=1}^{p}(N_{ij}+N_{ik})}} \]
\[ BC_{jk}=\frac{\sum _{i=1}^{p}(N_{ij}+N_{ik})-\sum _{i=1}^{p}2\;min(N_{ij},N_{ik})}{\sum _{i=1}^{p}(N_{ij}+N_{ik})} \]
\[ BC_{jk}=\frac{\sum _{i=1}^{p}(N_{ij} - min(N_{ij},N_{ik}) + (N_{ik} - min(N_{ij},N_{ik}))}{\sum _{i=1}^{p}(N_{ij}+N_{ik})} \]
\[ BC_{jk}=\frac{\sum _{i=1}^{p}|N_{ij} - N_{ik}|}{\sum _{i=1}^{p}N_{ij}+\sum _{i=1}^{p}N_{ik}} \]
\[ BC_{jk}=\frac{\sum _{i=1}^{p}|N_{ij} - N_{ik}|}{1+1} \]
\[ BC_{jk}=\frac{1}{2}\sum _{i=1}^{p}|N_{ij} - N_{ik}| \]
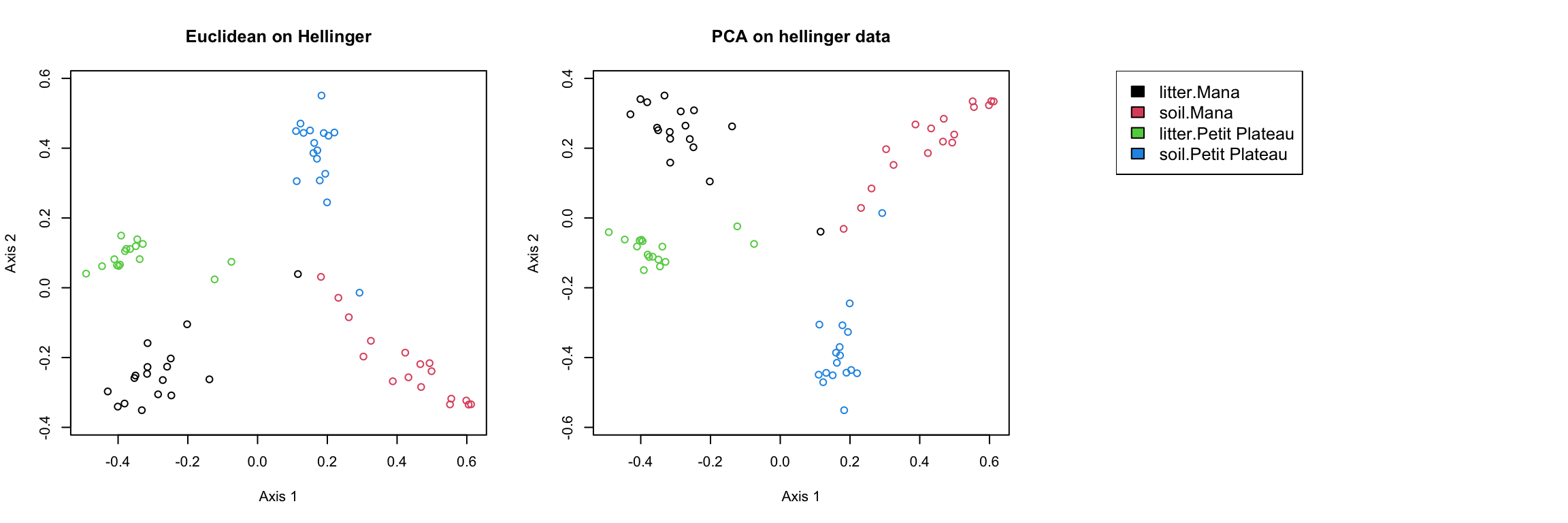
Principale coordinate analysis (1)
guiana.bc.pcoa = cmdscale(guiana.bc.dist,k=3,eig = TRUE)
guiana.euc.pcoa = cmdscale(guiana.euc.dist,k=3,eig = TRUE)
guiana.jac.1.pcoa = cmdscale(guiana.jac.1.dist,k=3,eig = TRUE)
guiana.jac.10.pcoa = cmdscale(guiana.jac.10.dist,k=3,eig = TRUE)
guiana.jac.50.pcoa = cmdscale(guiana.jac.50.dist,k=3,eig = TRUE)Principale coordinate analysis (2)
Principale composante analysis
Comparing diversity of the environments