| Home | Trees | Indices | Help |
|
|---|
|
|

|
|||
|
|||
|
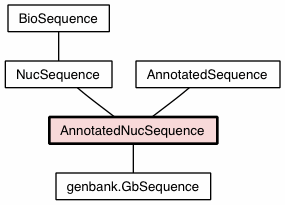
Inherited from Inherited from Inherited from Inherited from Inherited from |
|||
|
|||
|
Inherited from Inherited from Inherited from |
|||
|
|||
|
| Home | Trees | Indices | Help |
|
|---|
| Generated by Epydoc 3.0beta1 on Tue Apr 29 13:03:49 2008 | http://epydoc.sourceforge.net |